The motivation and theory for Ecotype Simulation may be found in our recent paper in PNAS by Koeppel et al.
Ecotype Simulation models the sequence diversity within a bacterial clade as the evolutionary result of net ecotype formation (here termed omega), periodic selection (termed sigma), and drift (termed d), yielding a certain number of ecotypes (termed npop).
Ecotype Simulation determines the rates omega, sigma and d, and most importantly, the number of putative ecotypes (npop) represented in an investigated set of sequences. Ecotype Simulation also allows demarcation of the sequences belonging to each ecotype. Here is an overview.
In the first step (Part 1, the Parameter Solution), the observed sequence diversity for the clade of interest is quantified as the “clade sequence diversity curve.” Then, using a brute-force method, Ecotype Simulation looks over a very broad space of parameters (including thousands of combinations omega, sigma, d, npop, over many orders of magnitude) for the most promising region of the space to yield the observed clade sequence diversity pattern with maximum likelihood. Next, the downhill simplex method (termed here “hillclimbing”) is used to more precisely identify the solution. In this first step, Ecotype Simulation also determines the 95% confidence interval of the parameter solutions.
In the second step (Part 2, the Demarcation), Ecotype Simulation demarcates the individual sequences belonging to each ecotype.
The program can accommodate at most 2000 sequences and 3000 nucleotides.
We will soon set up an on-line registration, but in the meantime we request that you e-mail Fred Cohan to let him know that you are an Ecotype Simulation user. We will then be able to send you updates of the program (and possibly fixes for as yet undiscovered problems).
Before you can run this program, you must have the latest version of java installed. You can do so by navigating to the java download page and downloading the latest update (you will need at least java 6 to get the program to run properly).
To run the ecotype simulation program, first download it. It is in the form of a self-extracting executable. To extract it double click on the executable and choose a directory to save it to.
You MUST also download NJPlot.exe from the NJPlot Website . Once at the website, click on MSWindows to download njplotWin95.exe. When it prompts you to extract, navigate to the folder that you extracted ParameterSolutions.exe into and extract NJPlot to the "Files" folder. Make SURE that njplot.exe is in the "Files" folder within the ParameterSolutions folder.
When you download the Ecotype Simulation program, you are also required to register on the Phylip website. Navigate to the Phylip Website
To run the ecotype simulation program, in the main ParameterSolutions folder click on the ParameterSolutions shortcut file (Fig. 1), which will start the program.

Two windows will open:
A) A DOS-window. In this, all the current calculations will
be displayed
B) A Java-window for opening and saving files and for
navigating through the ES software (Fig. 2).
· To run the ecotype simulation, first open up a fasta file
in the JAVA Window by going to
File->Open Fasta File
The fasta file must be formatted such that lines alternate
with >genename and then the entire gene sequence on the
next line. If the sequence for each gene is not on a single
line, the program will not function correctly. If your
sequences are in blocks, they can be converted to a single
line format using BioEdit™ or other sequence-editing
software.
Ø It is highly recommended not to run more than 200 sequences at a time
through this simulation. If you have more than 200 sequences, you may
consider randomly sampling from your sequences until you get down to under
200 sequences. Another possibility is to divide your clades into smaller
subclades that each contain 200 or fewer sequences, and then analyze each
subclade separately. 2000 sequences is the maximum, regardless of
the amount of time you're able to invest in analysis.
Depending on the speed of your computer, running 200
sequences may take a week or more.
At present, each sequence can be as long as 3000 nucleotides.
Ø Note that the fasta input file must contain an outgroup, and that the outgroup must be the first sequence in the file. If you intend only to get the parameter solutions without demarcating ecotypes (see below), then an outgroup is not strictly necessary, but the program expects an outgroup and so automatically ignores the top sequence in the file. So you should add a placeholder sequence at the top of your fasta file if you do not have an outgroup.
· To automatically get parameter values for omega, sigma, npop, and drift, click “Run through hillclimbing.” Be advised that this may take a long time, up to several days depending on the number of sequences and the speed of your computer.
· Many combinations of parameter values are evaluated for the fraction of replicate runs that yield a close fit to the observed clade sequence diversity curve (e.g., as seen in Figure 1 of Koeppel et al.; this curve represents, for each of a series of sequence identity criteria, the number of sequence-identity bins required to encompass all the sequences from the clade; also see description below “what the Parameter Solution software actually does”). For a given replicate run, a successfully close fit to the observed diversity curve is defined as being within a chosen “precision factor.” For example, in our analyses of Bacillus simplex, a replicate run was deemed successful if the simulation’s number of bins for each sequence identity criterion was within a factor of 1.5 of the observed value.
Ø The user may allow the program to choose the precision factor by selecting the “no criterion” option. The program will then choose the most precise factor that can be run through hillclimbing in under 8 hours. Otherwise, the program will attempt to use a user-selected factor.
Ø If you prefer to choose a precision factor, click on the drag-down menu under “select criterion” and choose the criterion that you want (5x, 2x, 1.5x, 1.25x, 1.10x, 1.05x).
· Once you have parameter values for a set of sequences, you can save a progress file by going to File->Save Current Project. The results will be saved in a *.cpm file.
· Next, you can run confidence intervals to find the 95% confidence interval for the parameter estimate.
Ø You can choose to run confidence intervals on none, any, or all of the following parameters: omega, sigma, npop, or drift.
· Alternatively, if you click on “Run Everything” the program will calculate both parameter values and confidence intervals for all parameters
· A narrative for the process of generating the parameter
solutions, which contains the input and output files for all
individual runs, is automatically saved by the program under
the name narrative.txt when the program is closed.
Copy and rename this file in order to have a record
of the analysis.
We strongly recommend that you read this section, as it will help you to use Ecotype Simulation with understanding and not as a black-box method.
A) The quantification of the observed sequence diversity by the clade sequence diversity curve
1. When you click open, it accepts a properly formatted, aligned fasta file (meaning the file alternates between >gene name on one line and then a gene sequence). In addition, this fasta file must include an outgroup as the first strain.
2. In the second step the file is copied (without outgroup)
to sequencesfasta.txt; the file numbers.dat is written and
formatted correctly with the gene length and number of
sequences; and then removegaps.exe, readsynec.exe,
correctpcr.exe, divergencematrix.exe, and binningdanny.exe
are all run. These programs will remove a nucleotide position
from all sequences if that position contains a gap in any
strain; they also “correct” a (usually small) number of
single substitutions based on the PCR error rate. Finally,
the sequences will be binned using a complete linkage
clustering algorithm (see the Fig. 3 below).
Ecotype Simulation quantifies the sequence diversity in the
observed sequence set by generating a clade sequence
diversity curve. For this, the sequence diversity is
clustered into bins at different sequence identity
criteria. For example, a bin at a given sequence identity
criterion, e.g., 95% sequence identity, is characterized
by encompassing only those sequences which have at least
a sequence identity of 95%.
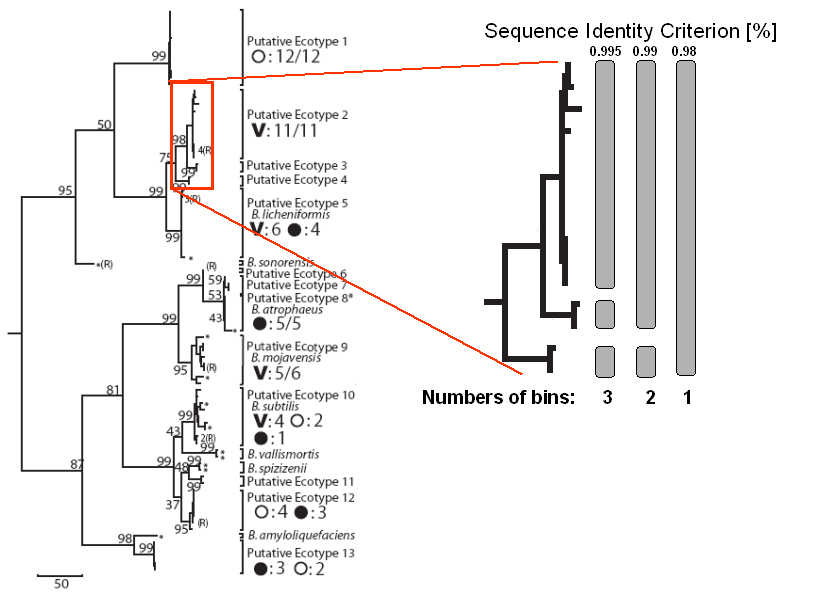
We exemplify this with the subclade of the Putative Ecotypes
2, 3, and 4 from the Bacillus subtilis-B.
licheniformis clade (Fig. 3a from the PNAS paper Koeppel
et al) (see below).
At a sequence identity criterion of 0.995 three different
bins are present, represented by Putative Ecotypes 2, 3, and 4. At a
sequence identity criterion of 0.99, two different bins are
present, represented by Putative Ecotype 4 and the pool of Putative
Ecotypes 2 and 3. At a sequence identity
criterion of 0.98 only one bin is present, encompassing
Putative Ecotypes 2-4. In that way, for a series of different sequence
identity criteria, the numbers of bins is determined and
graphically represented.
As a result, the clade sequence divergence curve is created (Fig. 4)
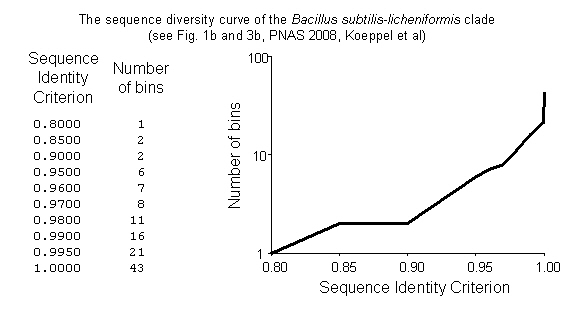
3. Next, the file “output.dat” which contains the binning data (i.e., the observed clade sequence diversity curve) is read in, the bins are written to the narrative file (narrative.txt – which will contain all the inputs and outputs to all programs as well as results), and then the bins are stored so that they can be used as additional input for the parameter estimation steps.
B) The brute-force method
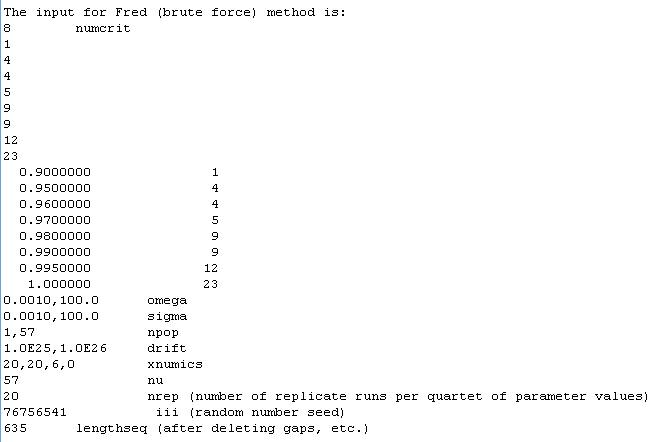
4. The next step is to run the brute-force method, where many parameter combinations, spanning many orders of magnitude, are evaluated for their ability to yield the observed clade sequence diversity curve. The input file is written automatically with the correct format (including numcrit, which is the number of sequence identity criteria, the binning data (representing the clade sequence diversity curve), omega and sigma ranging from 0.001 to 100, npop ranging from 1 to the number of sequences observed, drift at 1.0e25, xnumincs, which is the number of increments for omega, sigma, npop, and drift, respectively, nu = the number of sequences, and the number of replicate runs per parameter combination (nreps), which is always set to 20 replicates. A random number seed is generated, and the length of the sequence is also written. The input data are shown below.
5. Next the brute-force method is run with the above input, and the program writes the output into acinas.out . Each parameter combination that yields at least one successful fit to the observed data, at the 5x precision factor, over the 20 replicates, is included in the output. For each such parameter combination, acinas.out contains the values of the four parameters (in order of omega, sigma, npop, and d), followed by the fraction of successful fits to the observed data at the following precision factors (and in this order): 5x, 2x, 1.5x, 1.25x, 1.1x, and 1.05x.
C) Identification of a promising region in the Parameter Solutions space
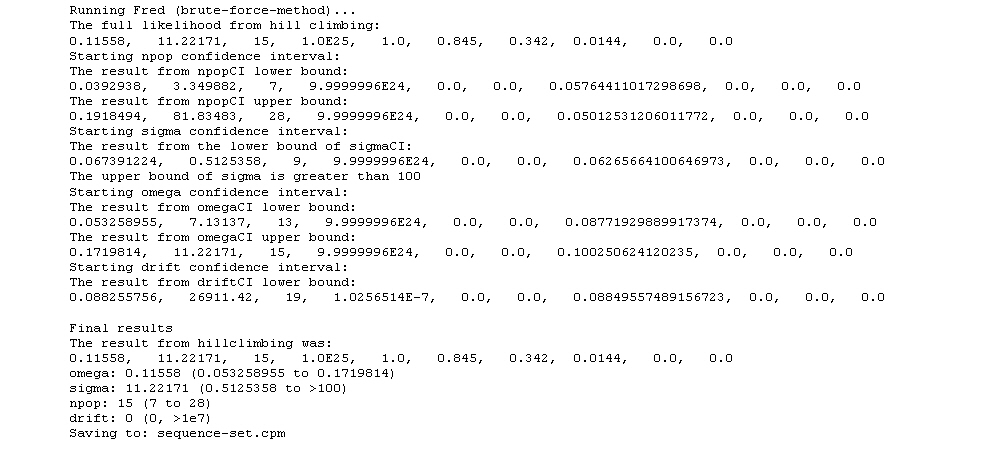
6. The results of the brute-force method are then sorted to yield the parameter solution to be used as the seed for the downhill-simplex approach (termed here as “hillclimbing”). From the output of the brute-force method, we identify the column of greatest precision that contains a solution with a likelihood value of 0.1 or greater (i.e., where 2/20 replicates scored a successful match to the observed clade sequence diversity curve). (We call this column X.) Then we sort the parameter solutions by the likelihood of successful match in column X. The sorted list is stored in fredDebug.dat in case there is a question as to the value chosen for hillclimbing.
D) The hillclimbing step (downhill simplex method)
7. The chosen parameter solution is then tested more fully (i.e., with 10000 replicates), so as to get a good estimate of the approximate likelihood of this solution producing a successful match to the observed data. The likelihood is calculated for each of the six precision values (i.e., 5x, 2x, etc.).
8. The program then estimates how long it will take to run the hillclimbing step. Starting with the precision level associated with column X, the program calculates how many replicates would be required to generate 50 successes, based on the 10000 replicates of the above step. Then, based on the amount of time it took to run the 10000 replicates of the above step, it calculates the time needed to assay one parameter combination for its likelihood value. Then, in anticipation of the hillclimbing step requiring 50 iterations of parameter combinations, it calculates the expected time to complete hillclimbing. If the hillclimbing is predicted to take longer than a preset number of hours (right now 8 hours) the program goes back, resorts by a less precise criterion, and then estimates the likelihood for that new parameter combination with 10000 replicates as before. It then checks the amount of run time again for hillclimbing. It continues this way until an appropriate value for hillclimbing is selected such that it will run in a reasonable amount of time.
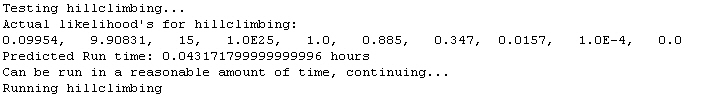
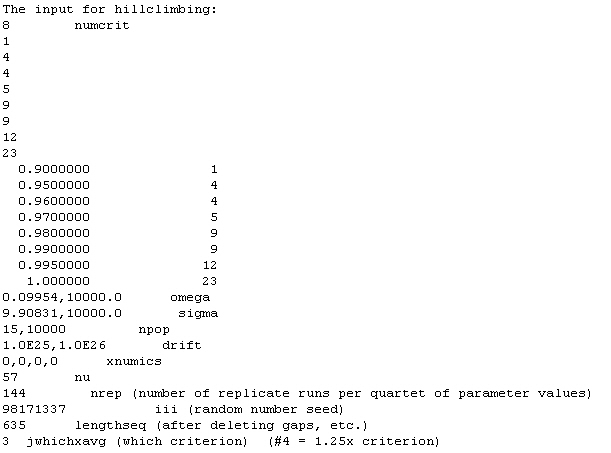
9. Hillclimbing is then run on the given value with enough replicates such that in the first iteration of hillclimbing, approximately 50 successful matches to the observed data are obtained. This value of nrep is obtained as 50 divided by the likelihood for the parameter solution.

E) Estimation of the 95% confidence intervalls
10. Next are the confidence intervals. Omega and sigma confidence intervals are run to a lower bound of 1e-7 and an upper bound of 100, with 6 increments per order of magnitude. If sigma runs up to 100 without getting out of confidence we just say that sigma exceeds 100. Drift is run from infinity down to 1e-7 with 3 increments per order of magnitude, and then the final value is inverted to get the actual drift value. The npop confidence interval is run down to 1 for the lower bound and up to the number of sequences for the upper bound. The last value still in confidence is reported in all cases. Additionally, the program aims for 20 successes at the threshold likelihood, ie for all two tailed tests (npop, sigma, omega) it is the likelihood from our hillclimbing parameters divided by 6.83, and for one tailed tests we divide by 3.87.
11. All the appropriate inputs and outputs are stored in narrative.txt as well as the final values from the confidence intervals and hillclimbing.
During the run of Parameter Solutions, all major intermediate steps and results are logged in the Java window. A typical log may look as follows:

· You can also run demarcations (separate strains into ecotypes) on the data by clicking on “other programs” and clicking on demarcations.
Ø Note: you can only do this after running through hillclimbing or after loading a saved file of data that has been run through hillclimbing
· Once in demarcations, you must load a fasta file again: go to file and click “open”
Ø Note that this fasta file must contain an outgroup, and that the outgroup must be the first sequence in the file.
Ø Once you select an appropriate file, you must choose to generate a Neighbor Joining or Parsimony Tree.
Ø Alternatively, if you already have a tree formatted correctly you can load that tree instead by going to File->Load Newick Tree File. You will then be prompted to load the proper fasta file as well
· Once you have loaded a fasta file the tree will pop up in NJPlot™ and the sequences will be listed in the demarcations program under “All Sequences.”
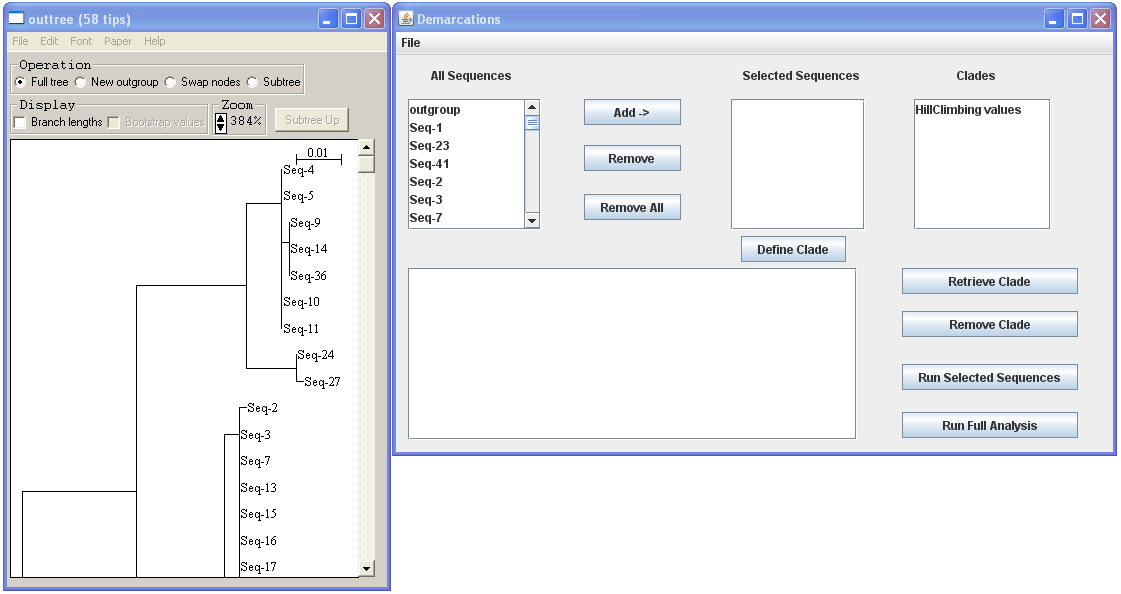
· You can select a group of sequences, click “Add” to add to select them, and then click “run selected sequences” to see if those sequences are consistent with being one ecotype. If they are, the program will say that the confidence interval included 1, meaning that the possibility that those strains represent a single ecotype is within the 95% confidence range.

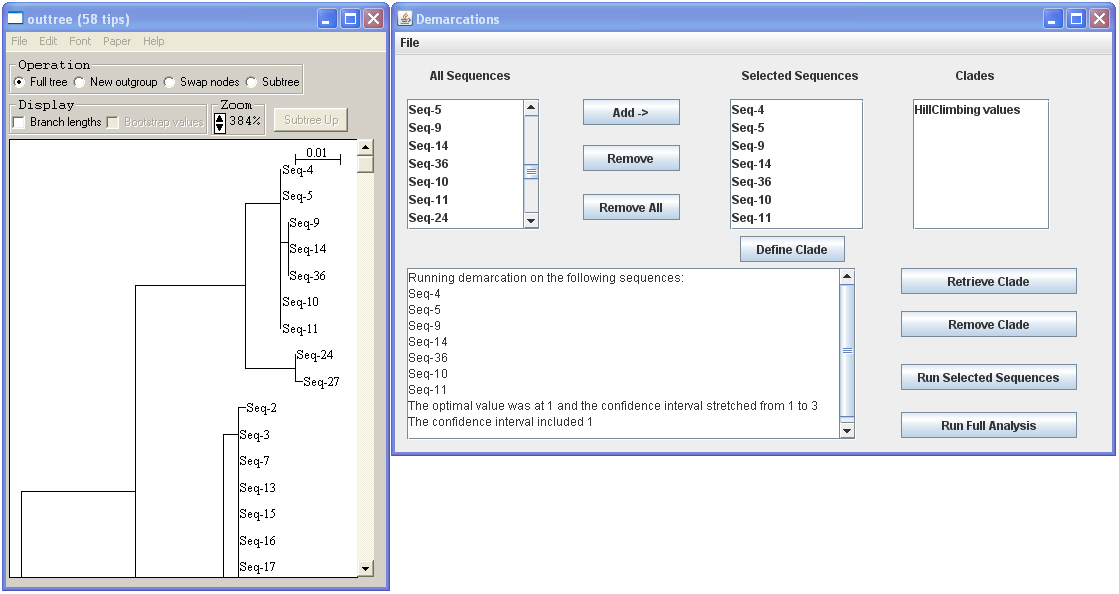
In this example above, all sequences in the observed cluster are consistent with being members of a single ecotype. Adding two more sequences (Seq-24 and Seq-27) will lead to a set of strains that are not anymore consistent with being a single ecotype (see the figure below).
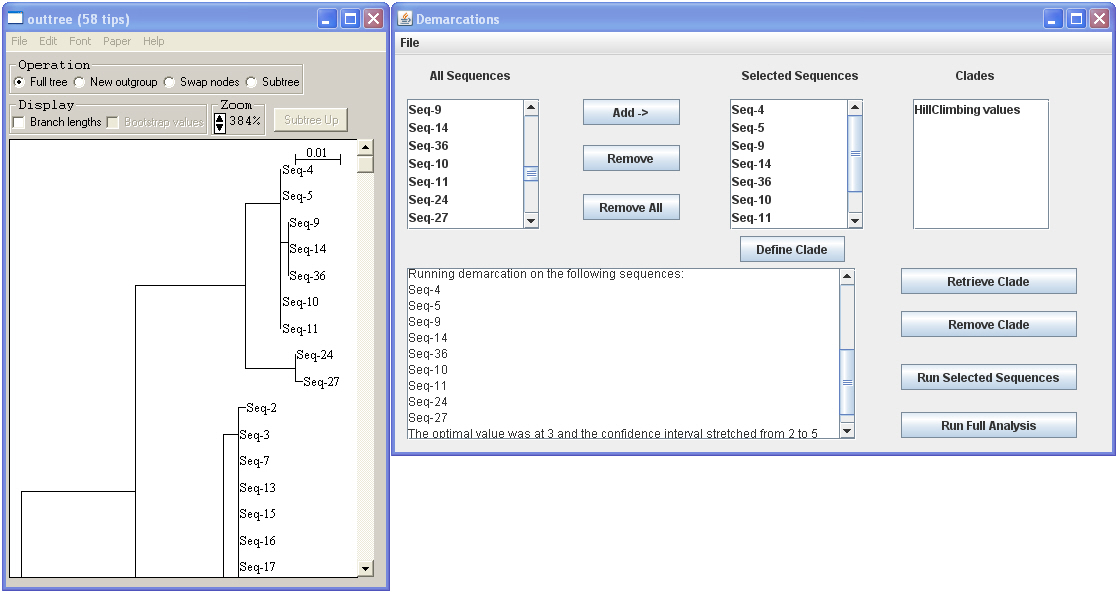
· You can continue to add and run sequences and then when you are satisfied that you have found the largest clade that is consistent with being a single ecotype, you can click “Define clade” to add them to the clade list. If you want to edit that clade again you can click on it and click “retrieve clade” to work on it. If desired, the sequences of a defined clade can be saved in their own separate fasta file, by retrieving the clade, and then selecting “Save Sequences as Fasta File” from the dropdown menu.
· A narrative for the demarcation process containing the
input and output files for all individual runs is
automatically saved by the program under the name
narrdemarc.txt when the program is
closed.
Copy and rename this file in order to have a record
of the demarcation analysis.
If you publish results from Ecotype Simulation, please cite our PNAS article:
Koeppel, A., Perry, E. B., Sikorski, J., Krizanc, D., Warner, W. A., Ward, D. M., Rooney, A. P., Brambilla, E., Connor, N., Ratcliff, R. M., Nevo, E. & Cohan, F. M. (2008). Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proceedings of the National Academy of Sciences 105, 2504-2509.
This work was supported by
NSF FIBR grant
EF-0328698
by NASA Exobiology Program grant NAG5-8824.
Updated February 20, 2008